On AAG serialization to a binary file
Motivation
Up until now, AAG could only be serialized as a JSON text file. In this article, we will talk about its binary serialization. Binary or not, any kind of AAG serialization in Analysis Situs is currently a one-way conversion. Here's the thing. AAG is inherently linked to a TopoDS_Shape because it caches B-rep elements in its internal hash maps. We demand that geometric information be removed during serialization and that just the pure graph be serialized. The motivations for this requirement are as follows:
- We are looking for a compact way to represent AAG serialized, while shape dumps, although possible, will make the serialized buffer explode in size.
- A serialized graph is often needed by a receiving system that is not concerned with native B-rep geometry as such. Examples include pattern-matching techniques and graph-based machine learning, to name a few. Therefore, there is often just no need to serialize geometries.
- There is, in theory, a deterministic procedure that would allow the serialized AAG to be reapplied to its originating CAD model. As a result, the serialization process does not inherently lose any data: we just need to have a subprogram to expand the serialized AAG back on a real piece of geometry.
When your CAD models are moderate in size, text files are totally fine. However, as a part's size increases, serialization to text formats becomes less efficient, even if no geometry is serialized. The underlying graph structure simply becomes too large. Assume that the attributes you specify in the AAG are "heavy," for example, you use the AAG of a part to carry over secondary shape descriptors, such as point clouds of sampled edges and faces. This could be relevant in machine learning contexts when switching from accurate B-rep geometry to a quantified shape descriptor that a neural network can understand. In these cases, binary serialization of the AAG with all of its features would aid in data communication from the rule-based feature recognition system (Analysis Situs) to another software package designed for AI-based recognition.
Serialization tools
Binary streaming is handled by a number of internal tools in OpenCascade:
- BinObjMgt_Persistent is a class for binary serialization in OCAF.
- BinTools_OStream is a geometry-oriented buffer used for saving binary BREP files.
- Common C++ functions for binary streaming are also seen throughout OpenCascade sources, for example, in the STL mesh reader.
- Plain C (non-C++) functions, such as fread() and fwrite() are also seen in the library, e.g. in STL binary writer (don't ask me why the reader and the writer employ different styles of working with files).
- There are some legacy interfaces, like FSD_BinaryFile which effectively wrap the lower-level C functions.
For AAG serialization, we use the standard C functions, not employing anything from the OpenCascade toolset. Personally, I find C functions way cleaner compared to C++, which, although powerful, tends to unreasonably complicate conceptually simple things.
Binary format highlights
In this section, we go over several format details. Please keep in mind that what follows is not a thorough format reference but rather a description of the format "ideology" that should help you understand how AAG might be serialized to communicate with third-party software.
Header and adjacency lists
First, let's look at the following HEX buffer, which is an example of a possible AAG serialization format (this is the HxD viewer):
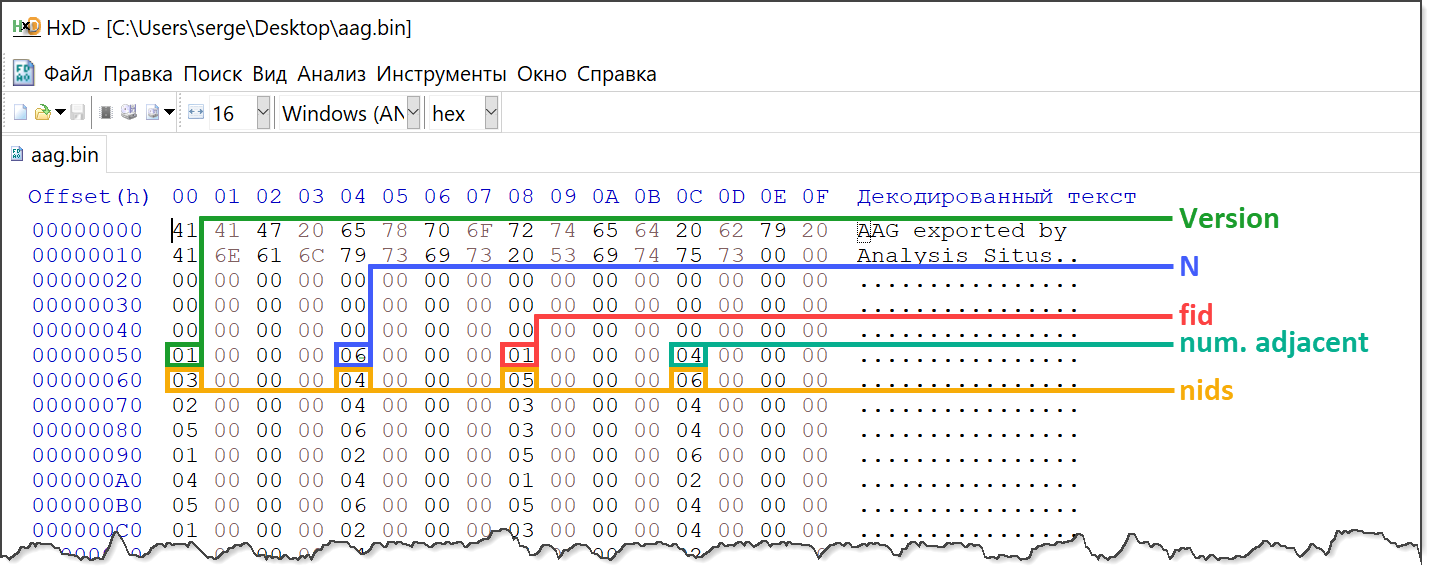 |
Here we have the following designations:
- Version is the binary format version (just one integer value);
- N is the number graph nodes (CAD faces);
- fid is the next 1-based face ID in the adjacency row;
- num. adjacent is the number of neighbors for fid;
- nids are the faces adjacent to fid.
It is quite obvious that such an adjacency matrix (with variable row lengths) defines an undirected face adjacency graph (FAG). In the "header" section, we reserve 80 bytes for something that will identify the binary format. We don't follow any "best practices" here for the sake of simplicity, but it might make sense to add a checksum and some magic numbers to the file header as well.
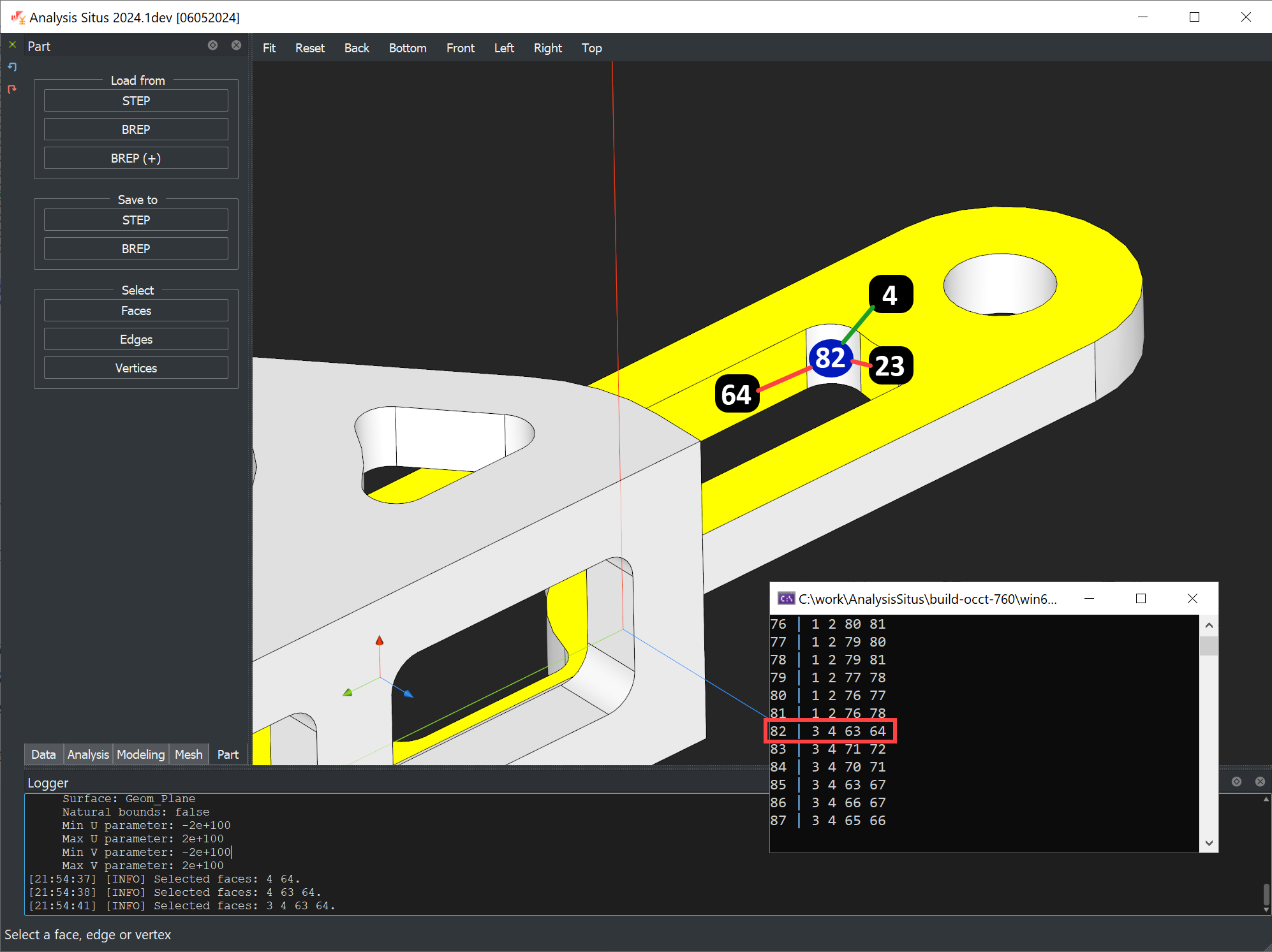
For a primitive box shape composed of 6 faces, the adjacency matrix stored in the binary buffer has the following data:
fid | nids ----+--------- 1 | 3 4 5 6 2 | 3 4 5 6 3 | 1 2 5 6 4 | 1 2 5 6 5 | 1 2 3 4 6 | 1 2 3 4
The adjacency information can easily be verified in the GUI of Analysis Situs. The stored data is not a two-dimensional array because we do not store zero elements to save space.
 |
Node attributes
On top of the serialized adjacency relations, we should now add the node and arc attributes, transforming the empty FAG data structure into a more feature-rich AAG. Every attribute can be serialized into its own data block, making it easy to locate it in the binary file. The objective is to preserve the binary format's backward and even forward compatibility by letting the application disregard any unknown attributes. Here is a format for a nodal attribute:
NODE_ATTR_BEGIN <fid> <attrName> <buffSize> //-//-// buffer //-//-//
Here we have the following designations:
- NODE_ATTR_BEGIN is the keyword indicating the beginning of the attribute block;
- fid is the 1-based face ID (node ID), where this attribute is attached;
- attrName is the attribute name (class name);
- buffSize is how many bytes the following buffer occupies;
- buffer is how the attribute serialized itself;
An attribute is asked to serialize itself by calling the virtual Serialize() function, whose implementation is empty by default. It is up to a specific attribute to define its own serialization format, given that the framework takes responsibility for dumping the attribute's serialized buffer after the NODE_ATTR_BEGIN keyword.
Arc attributes
Arc attributes can be serialized almost identically to node attributes. Of course, instead of addressing faces by their 1-based IDs, we now need to make references to the graph links. Therefore, the format of the arc block looks slightly different:
ARC_ATTR_BEGIN <fid1> <fid2> <attrName> <buffSize> //-//-// buffer //-//-//
Usage of the binary format
By default, all AAG attributes come with empty implementations for serialization functions, so you, as a developer, are responsible for defining the way your custom data is turned into persistent bytes. This is no different from OCAF, where you're supposed to implement the so-called storage and retrieval "drivers." To make things work, the dedicated Serialize() virtual method should be implemented for each attribute you want to have in the binary file. Here's the signature:
virtual bool Serialize(FILE* pFile) const
{
(void) pFile;
return false;
}
This method accepts the FILE handler of the binary file, which is assumed to be opened. You can then use the standard fwrite() C function to get the job done. If the attributes do not provide any serialization logic, they are still dumped to the binary file with empty buffers. This way, you at least know that the attributes are present in the graph.
 |
As mentioned in the beginning, our main goal for AAG serialization was to make a bridge between rule-based feature recognition and deep learning (with DGL). It required that the produced binary file be interpreted in the corresponding Python code, where we needed to implement the corresponding reader. Luckily, working with binary files in Python is quite straightforward. Here's a code excerpt to read the adjacency information (FAG) from the header of a binary file:
def readGraph():
graph = nx.DiGraph()
f = open(<filename>, mode="rb")
header = f.read(80)
numNodes = int.from_bytes( f.read(4), 'little' )
print('We have %d nodes in the FAG.' % numNodes)
# Create FAG without attributes
nodeCounter = 0
while nodeCounter < numNodes:
nodeCounter += 1
fid = int.from_bytes( f.read(4), 'little' ) # Next face ID
numAdjacent = int.from_bytes( f.read(4), 'little' ) # Num. of adjacent faces
# Read adjacent
adjCounter = 0
while adjCounter < numAdjacent:
adjCounter += 1
nid = int.from_bytes( f.read(4), 'little' ) # Next face ID
graph.add_edge(fid - 1, nid - 1) # Graph nodes are 0-based in Python
f.close()
The complete example is published as readBinaryGraph.py script in the repository of Analysis Situs.
Want to discuss this? Jump in to our forum.